About rings and strings: novel principles in the organization of chromatin
The complexity of storing, distributing and accessing DNA
Each single nucleus of our body cells contains 46 DNA-molecules with a total length of about 2 meters. These extremely long molecules not only have to be packaged to fit into the small nuclei. They have to be copied and distributed evenly during cell division and, above all, they have to be accessed permanently. Accessibility of specific DNA-regions, their vicinity or distance to specific enhancing regions determines to which extent a certain element of genetic information is active in a certain cell. Cohesins are proteins forming ring-like structures and have been shown to be involved in several of the processes summarized above.
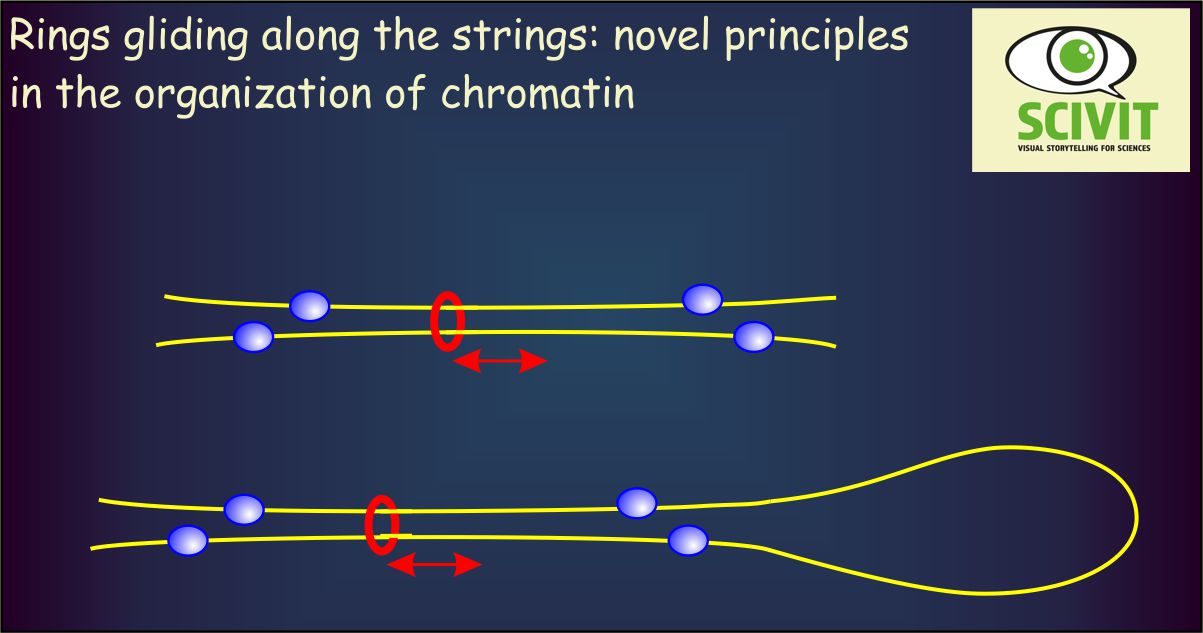
Cohesins: ring-like structures inclosing DNA-strings
Cohesin-rings are assumed to bind two different DNA-strings together, this way either connecting to different DNA-strands or forming loops of one strand. How are the DNA-regions inclosed by cohesins defined and how flexible and dynamic are cohesin-DNA-interactions? Davidson and colleagues used single-molecule imaging to study the precise actions of cohesins when binding to DNA. They demonstrated that cohesin-rings can indeed inclose DNA-strings and move along such strings rapidly. Movement of rings is blocked, however, by transcription and by proteins binding to specific DNA-sequences (CCCTC). This way, a given cohesin ring can be fixed within a given region of the DNA-string, consistent with its assumed function in physically connecting two different DNA-regions.
Neueste Kommentare