Bacterial metabolism is coordinated in a simple way
Bacterial metabolism comprises numerous enzymes, genes and regulating factors
Even small, simple bacteria possess a complex metabolism depending on numerous different enzymes. Each enzyme is encoded by one or several genes, controlled by one or several transcription factors, which on their turn may be activated or deactivated by various factors. How is all this coordinated?
How can metabolic regulation be assessed?
Kochanowski and colleagues presented a comprehensive analysis of metabolic regulation in the bacterial model organism Escherichia coli. They imposed 26 different growing conditions on their model organism. Using fluorescent reporter proteins they studied the activity of 95 different genes under these conditions. In parallel they analyzed metabolite levels under experimental conditions and searched for metabolites correlating with gene activity.
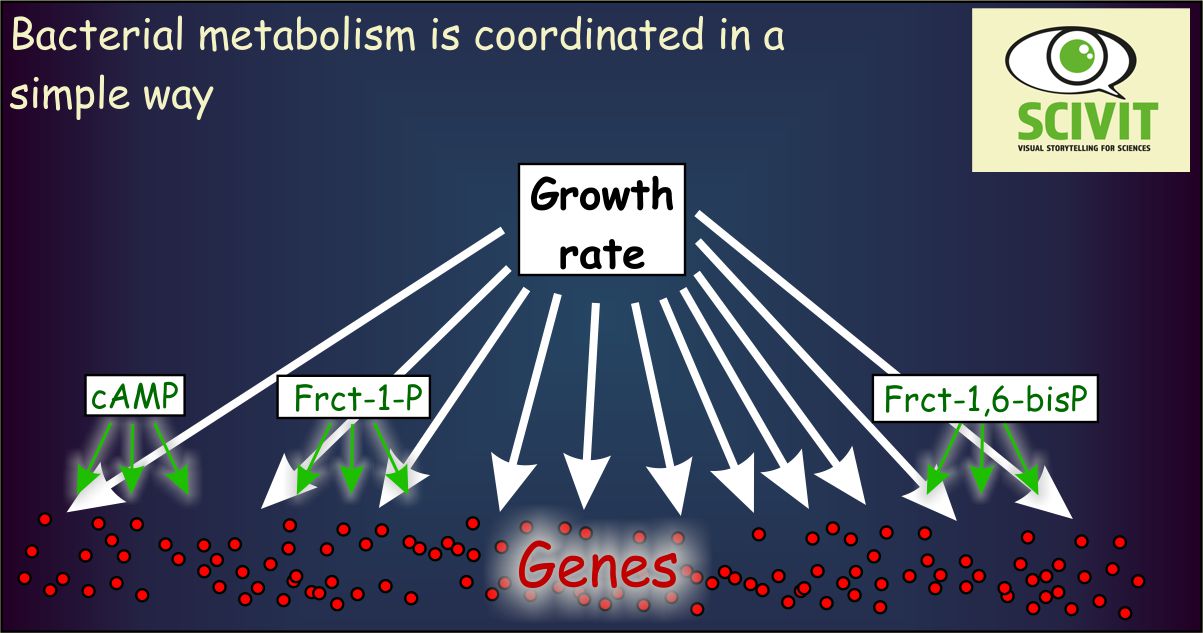
There are only few metabolic regulators in Escherichia coli
Most of the genes analyzed showed very similar patterns of activity correlating with bacterial growth rates. There were only few gene promoters activated in a specific way depending on actual metabolic conditions. And there were only few metabolites correlating with this specific activity. Cyclic AMP, fructose-1,6-bisphophate and fructose-1-phosphate apparently are sufficient to adapt metabolic activity of Escherichia coli to all environmental conditions encountered in the experiment.
Neueste Kommentare