Recently I became aware of a bug in the virtual genomic planets which I presented some posts ago. This post will be about my way of fixing this bug and I hope there will be some comments improving my approach even further. So this is rather a technical post…
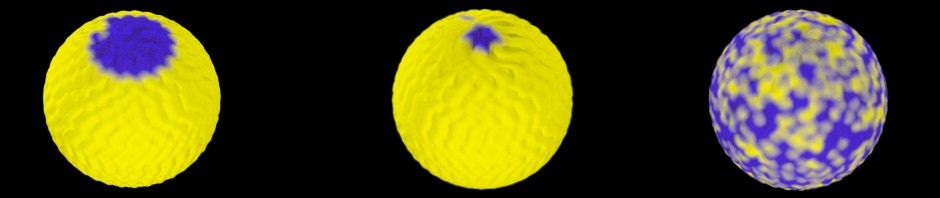
The genomic planets often contained mountain ranges in the polar regions and oceans at the equator. This was somewhat odd, but at least in some cases it correlated with the positions of centromers. In addition, I was sure that the distribution of genes on the sphere was correct, i.e., that the arc separating two genes really corresponded to the genomic distance of these genes. This is, however, not sufficient to ensure a realistic distribution of oceans and continents, as I had to realize. To examine this distribution, I constructed a planet from an artificial genome of equidistant genes, the size of the Yersinia pestis genome. In this planet regions with gene distances below a certain threshold are depicted as blue mountain ranges, the rest is colored in yellow. Since the artificial genome is composed of equidistant genes, the whole planet should show a uniform land type. This is, however, as shown in the image below, not the case.
- Test planet for equidistant genome
As observed in many “real” planets, even this homogenous planet contains mountain ranges in the polar regions. It took me a while to realize the reason. First I checked once again, whether the genes constituting the planet are really separated by identical arcs. They are as you can see below for the same planet , where gene positions are given as small blue or yellow dots (only the northern hemisphere is depicted here).
- Gene distribution in equidistant planet
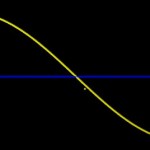
The planet surface is calculated from the actual distance of the points representing genes, not from the respective arc. This actual distance is always smaller than the arc with the error depending on the respective angle. With equidistant genes the angle decreases with latitude. In consequence, the large angles from the polar areas result in larger errors and smaller actual distances than the small angles around the equator. How to cope with this problem? I was not able to find a way to use distances instead of arcs for the planetary distribution of genes. The only way I could figure out was to distort gene distribution in a way reducing the densities of genes around the pole and increasing it at the equator. For this purpose I added the following function to the gene distribution. (Actually I applied three different functions covering three different regions of the genome, but I don’t want to go too much into details…)
- Function for correcting the gene distribution
The function runs over the complete genome and every gene position is increased or decreased by its respective value. When this correction is applied the equidistant genome produces a more homogenous planet.
- Corrected Planet
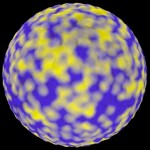
This is not the last word, however, since I became aware of another, better fix: I found a way of using the arc separating two genes rather than their distance for calculating surface features. The application of this approach to the artificial genome leads to uniform planets. Both area features occur in such planets only when the threshold between these features is set to a specific value. In this case yellow and blue areas are distributed randomly, which demonstrates that the new way of calculation is indeed unbiased.
Further reading:
Genome may be mostly junk after all
Now that’s a f***ing big genome!
German language:
1000 Genome sequenziert und immer noch nichts passiert